
By Anurag S. Rathore, Rozaleen Dash
A major advantage of SPR-based analysis is its ability to estimate the association and dissociation rate constants, an advancement over the traditional steady-state analysis of biomolecules.
Biotherapeutics are an emerging class of treatment modalities that are manufactured using living cells. They have been successfully used for treating many life-threatening and chronic diseases. Compared to the traditional small-molecule (pharmaceutical) drugs, biotherapeutics are complex and have the ability to bind to more than one target molecule. Biosimilars provide a more affordable treatment option, and this is likely to become more relevant in the future as the affordability of these products remains critically poor in emerging and underdeveloped economies (1–3). The biopharmaceutical industry faces an increasing demand to accelerate process development while saving on cost and time. A possible solution to alleviate this is by using high-throughput potential tools such as surface plasmon resonance (SPR) to measure biomolecular interactions in real-time and in a label-free environment (4, 5).
Biomolecular interaction analysis (BIA) can be used in the following ways (3):
- To monitor the level of interactions between two or more species
- To determine the affinity of the interactions
- To estimate the actual association and dissociation rates
- To measure the concentration of one of the species.
SPR is a rapidly emerging tool for studying ligand binding interactions with membrane proteins, which are the major molecular targets for biopharmaceutical products (6). In SPR, one of the interactants is immobilized to the sensor surface and the other is kept free in solution and passed over the surface. Association and dissociation are measured and displayed in the form of a sensorgram. In this 41st article in the Elements of Biopharmaceutical Production, the authors present the basics of SPR as well as the various applications it offers in biopharmaceutical analysis.
Various measurements possible with SPR
Detection of specificity is comparably easy to perform in SPR. An analyte is injected and, after a certain amount of time, the response is measured. Plotting the response curve against sample number indicates the relative strength of binding. Bulk response caused by the medium in which the analyte is dissolved is the major drawback but can be easily avoided with proper reference measurements (7, 8).
Analyte concentrations are measured at very high ligand densities on the sensor surfaces. The rate of the binding constant is proportional to the concentration of the analytes. After a standard curve has been created, unknown samples can be measured quickly.
Equilibrium analysis is used to determine the strength of the binding. The first type is performed by using several analyte concentrations, which involve flowing the analyte over the ligand until the signal levels out and the net association is equal to the dissociation. By plotting the maximum response versus the analyte concentration, a line can be fitted to estimate the affinity constant (KD). The second type of experiment involves putting the two interactants together and incubating them until equilibrium. One of the interactants is to be constant, and the other is varied over a range of concentration. The concentration of the free analyte can then be determined after equilibrium.
Kinetic rate analysis is used to investigate the behavior of the system. Interaction kinetics describes the interaction between one or more components. After the interaction, the components leave each other unchanged as opposed to enzyme kinetics. The rate of association is determined in real time when the analyte flows over the ligand. Over time, the buffer replaces the analyte and the dissociation rate of the analyte is monitored. Both the association and dissociation curve can be fitted to one of the chosen models. In addition, the equilibrium constant can be calculated (9, 10). There are two types of experimental approaches available for kinetics experiment: multi-cycle kinetics (classical “standard” approach) and single-cycle kinetics (see Table I).
Various techniques are used to study structure-function affinity. The function is measured in terms of specificity (affinity), rate and equilibrium constants, as well as thermodynamic properties. While with the majority of SPR experiments the interaction conditions are held constant, varying these conditions (e.g., the temperature) can reveal important thermodynamic properties. SPR systems are capable of measuring the specific ligand–analyte interaction in real-time, which enables the researcher to simultaneously estimate both the rate and equilibrium constants (11, 12).
Calibration free concentration analysis (CFCA) has been developed for measuring the active concentration of a ligand without a calibration curve (9). The method makes use of the mass transport limitation, which occurs when high-density ligand surfaces are used. By injecting the analyte at two different flow rates (e.g., 10–90 µl min-1), the active analyte concentration can be calculated from the slopes of the curves (13).
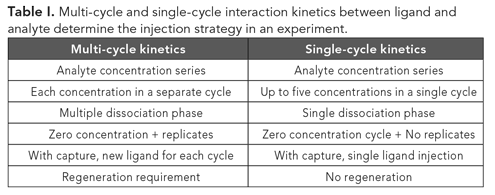
How much ligand to immobilize?
Table II shows the ligand immobilization on the sensor chip are as follows:
- Kinetics should be done with the lowest ligand density, which gives a good response without being disturbed by secondary factors such as mass transfer or steric hindrance.
- For specificity measurements, almost any ligand density will do as long as it gives a good signal.
- Concentration measurements need the highest ligand density to facilitate mass transfer limitation.
- Affinity ranking can be done with low to moderate density sensor chips.
- Low molecular mass binding should be done with high-density sensor chips to bind as much as possible of the analyte to gain proper signal.
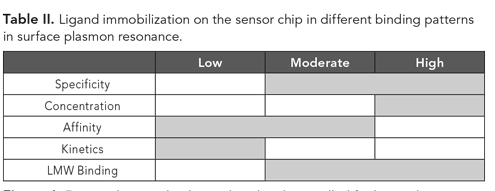
Analytical characterization and comparability of biotherapeutics
Biotherapeutics are complex products and, therefore, require characterization using numerous orthogonal analytical tools. Biosimilars undergo limited clinical examination prior to approval under the premise that comparability to the corresponding innovator product using an exhaustive analytical characterization exercise has been demonstrated (14). The comparability exercise involves characterization and analysis by a platform consisting of numerous, orthogonal analytical tools (Figure 1). Failing to demonstrate similarity can come at a significant cost and can trigger a more extensive (and expensive) clinical examination prior to receiving regulatory approval. Guidance documents from regulatory health authorities, such as FDA and the European Medicines Agency (EMA), emphasize the importance of extensive analytical characterization in showing the similarity between a biosimilar and its reference product based on a comprehensive assessment of protein structure and function (15, 16). As such, developers of biosimilars must put together a toolbox of state-of-the-art technologies. Companies such as Sandoz and Celltrion, which have both received FDA approvals for biosimilars, have had success in using real-time, label-free biophysical analytics as a platform approach for product and process support, evaluating target and receptor binding, and for measuring immunogenicity and specificity. Another significant challenge with biosimilars as well as other biological products is that of standardizing the Fc receptor binding. Fc receptors can be activating, or inhibitory, or without any effect in antibody-dependent cellular cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC). Characterizing these highly complex products and their interactions, however, is extremely challenging (13).
Figure 1. Factors that need to be analyzed and controlled for biosimilar development. (Figures courtesy of the authors)
Monoclonal antibodies (mAbs) are at present the fastest growing subclass of biopharmaceuticals and have been successfully demonstrated to treat a variety of diseases, mainly in oncology and auto-immune and infectious disease segment. Antibodies recognize their antigen through the variable regions of the antigen-binding portion (Fab). As a result, they may interfere with one or several functions of this antigen, leading to the therapeutic effect. On the other hand, through the constant regions (Fc), they may interact with Fc-binding molecules and recruit patient immune effector function to destroy the marked target. The ADCC is triggered by an interaction between the Fc region of an antibody bound to, for example, a tumor cell and the Fcγ receptors on immune effector cells, leading to the elimination of the tumor cell by phagocytosis or lysis, depending on the type of mediating effector cell. CDC is initiated by complement component C1q binding to the Fc region of the antibody, triggering activation of the complement that leads to cell death by phagocytosis, lysis, or disruption of the cell membrane (16). SPR is a well-established technique for detection and monitoring of biomolecular interactions in real time. It has been formatted as a parallel line assay and researchers have demonstrated high levels of accuracy, precision, and linearity with the assay, making it useful for establishing comparability, estimating potency and examining stability (17–20). SPR has also shown greater precision and reproducibility than the traditional cell-based assays such as ADCC and CDC.
SPR applications are not only limited to ligand–receptor interaction kinetics dynamic analyses, but they are also used for drug discovery and development. There are several different formats of SPR biosensors, including the array format, multi-channel unit format, and SPR imaging format, which allow simultaneous and continuous detection to analyze the performance of hundreds to thousands of affinity binding events on a chip surface. In SPR imaging, the incidence angle remains fixed and the binding of biomolecules on a gold surface is measured as the change in reflectivity in relation to the incident ray intensity, unlike SPR sensors that depend on the measurement of the absorption dip in the SPR angle or SPR wavelength. Despite the excellent benefits inherent in SPR technology, conventional propagating SPR biosensors have a serious limitation due to their inability to support multiplex analysis, as less than four analyses with conventional SPR instruments make such parallel operations feasible. In contrast, SPR imaging technology uses a multi-analyte biosensor that permits a high-throughput approach and achieves a similar degree of sensitivity that is achieved by conventional SPR biosensors. Therefore, SPR imaging systems without any labeling requirements are more suitable for high-throughput screening (HTS), particularly in drug discovery, than any other optics-based detection techniques (21, 22).
Antibody applications of SPR broadly fall into two categories, such as screening and characterization. Antibody screening often involves capture of an antibody followed by a single injection of antigen. The kinetics of binding can be estimated directly from a screening experiment and is of major interest for antibodies intended for diagnostic or therapeutic purposes. Antibodies with slow off rates are often selected as candidates to provide prolonged drug residence time and also potentially higher efficacy (23). A complicating factor in the selection procedures is that antigen binding may be heterogeneous, and such antibodies are often deselected in favor of monophasic binders that may be more specific. The selection is, therefore, focused not only on stable binding but also on monophasic binders. In antibody characterization, SPR is typically used for epitope binding, specificity, concentration, and kinetic/affinity analysis. When using SPR for establishing analytical comparability, the biosimilar product is compared with the reference product.
Binding patterns of SPR
Antibodies, cytokines, and hormones typically interact with their receptors. While cytokines and hormones retain their natural sequence and folding, antibody therapeutics are engineered to interact with target molecules (including antigens, Fc receptors, and complement factors) based on their intended mechanism of action. SPR data experiments were performed using SPR based sensor system (Cytiva) with analysis temperature set to 25 °C. Series S Sensor Chip CM5, Series S Sensor Chip NTA, Series S Sensor Chip Protein A HBS-EP buffer (10 mM Hepes, 0.15 M NaCl, 3 mM ethylene diamine tetra acetic acid [EDTA], and 0.05% [v/v] surfactant P20, pH 7.4), Amine Coupling Kit, Human Fab Capture Kit, NTA Reagent Kit, Protein L, and Biotin Capture Kit were all obtained from Cytiva.
Case study I: Binding kinetics of other rituximab biosimilars and Ristova to human FcRγIIIa (CD16a)
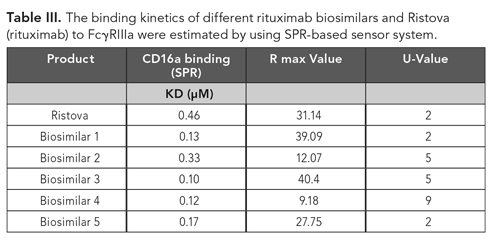
The binding kinetics of Ristova, a rituximab biosimilar, and other rituximab biosimilars to human FcRγIIIa (CD16a) receptor (R&D systems) were determined by SPR-based sensor system (Cytiva). Kinetic analysis of FcRγIIIa (CD16a) binding was performed by injecting different known concentrations of aggregates (from 0.25 to 4 µM) onto immobilized carboxymethyl dextran-coated CM5 sensor chips, which were used with His-coupling chemistry (13). All measurements were performed at 25 oC with a flow rate of 30 µL/min using HBS-EP buffer with association time 60 s followed by 60 s dissociation phase. KD were calculated from the sensorgrams using the 1:1 fit model using SPR-based sensor evaluation software (Cytiva) (Figure 2).
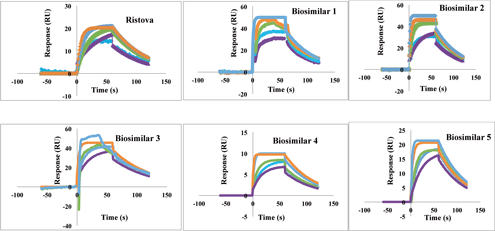
Figure 2. Binding kinetics of different rituximab biosimilars and Ristova (rituximab) to human FcγRIIIa (CD16a). The color key (-4 µM, -2 µM, -1 µM, -0.5 µM, -0.25 µM) is same for all the sensorgram indicates known concentrations of analytes.
SPR was used to provide a comparison of the binding kinetics of rituximab biosimilars to human FcγRIIIa with respect to Ristova. As given in Table III, the KD value of Rituximab biosimilars to FcγRIIIa was calculated to be within the same order of magnitude as Ristova. KD values were compared, wherein Ristova exhibited higher binding affinity than the other rituximab biosimilars. Biosimilar two exhibited comparable affinity for binding to FcγRIIIa with respect to Ristova, but others yielded lower values.
Case study: II Binding kinetics of GCS-F biosimilars and Neupogen (filgrastim) to GCS-F-R (CD114)
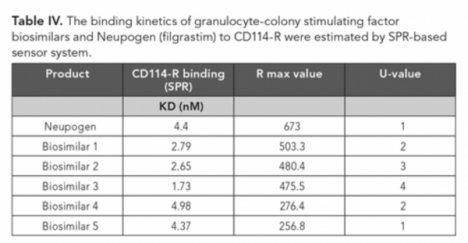
The binding kinetic interactions of five different biosimilars samples of granulocyte-colony stimulating factor (GCSF) to human CD114-R receptor (R&D systems) was determined by Biacore X100 plus biosensor (Cytiva). For GCSF-R affinity analysis, CD114-R receptor was immobilized on a CM5 sensor chip surface according to the manufacturer’s recommendation with a level of ∼200 response units (RU) reached. Samples were injected in a series of concentrations ranging from 2–32 nM (Figure 3) with association time 120s followed by 120s dissociation phase. All measurements were performed at 25 oC with a flow rate of 30 µL/min using HBS-EP buffer according to the manufacturer’s protocol. Kinetic constants were calculated from the sensorgrams using the 1:1 fit model using SPR-based sensor evaluation software (Cytiva) (Figure 3).
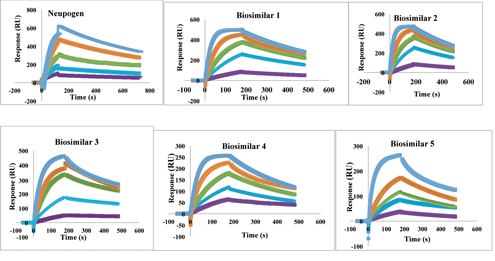
Figure 3. Comparative analysis of granulocyte-colony stimulating factor biosimilars and Neupogen (filgrastim) for affinity to CD114-R by surface plasmon resonance. The color key (-4 µM, -2 µM, -1 µM, -0.5 µM, -0.25 µM) is same for all the sensorgram indicates known concentrations of analytes.
Table IV represents the KD value of GCSF biosimilars to CD114-R was calculated to be within the same order of magnitude as Neupogen. Biosimilar four exhibited little higher affinities than Neupogen for binding to CD114R, but others yielded lower values. Because nearly all mAb biosimilars are produced in mammalian cells, they exhibit different post translational modifications (PTMs), which results in heterogeneity. This could be attributed to differences in expression system, culture conditions, purification processes, formulations, or storage conditions of the product and need to be suitably addressed as per the “similar but not identical” paradigm.
Data analysis: Key outcomes
The equilibrium constant determines the ratio of the antibody association rate (kon) of the antibody, how quickly it binds to its antigen, and antibody dissociation rate (koff), how quickly it dissociates from its antigen. Chi2 and residual values were used to evaluate the quality of fit between the experimental data and individual binding models. Plots of residuals indicate the difference between the experimental and reference data for each point in the fit. The Chi2 value represents the sum of squared differences between the experimental data and reference data at each point. Lower Chi2 values indicate a better fit; however, it is difficult to recommend absolute values for acceptance limits for Chi2. U-value indicates the uniqueness value for the kinetic rate constants (19). Lower values indicate greater confidence in the results. Prior to analysis, sensorgrams are double referenced by first subtracting data from a reference flow cell and then subtracting a blank cycle where the buffer is injected instead of protein sample. Sensorgram comparison requires that standards, controls, and samples have been generated in the same way using identical association and dissociation times (20, 21). The assay format must be the same, and it is not possible to mix multi-cycle kinetic data and single-cycle kinetic data in the same session.
SPR provides a meaningful insight into the characterization of biotherapeutics and comparability of biosimilars present in the market.
Conclusion
This review has demonstrated that SPR is a rapidly developing technique typically used for characterization of protein interactions and in screening for selection of antibodies or small molecules with preferred binding properties. In characterization, complete binding curves are normally fitted to defined interaction models to provide affinity and rate constants, whereas report points indicative of binding and stability of binding are often used for analysis of screening data. It is a high-throughput technique that can potentially serve as a useful tool to evaluate the higher-order structural integrity of proteins for characterization as well as comparability purposes. As an outcome of this review, SPR provides a meaningful insight into the characterization of biotherapeutics and comparability of biosimilars present in the market.
Acknowledgements
This work was funded by the Center of Excellence for Biopharmaceutical Technology grant under Department of Biotechnology, Government of India.
References
1. R. Karlsson, “Surface Plasmon Resonance in Binding Site, Kinetic, and Concentration Analyses,” in The Immunoassay Handbook, D. Wild, Ed. (Elsevier, San Diego, CA, Fourth ed., 2013), pp. 209-222.
2. A.S. Rathore, Trends Biotechnol. 27, 546–553 (2009).
3. G. Walsh, Nat Biotechnol. 32 (10) 992–1000 (2014).
4. D. L. M. Rupert, et al., Analytical Chemistry 88, 9980–9988 (2016).
5. O. K. Kari, et al., Drug Delivery and Translational Research 7, 228–240 (2017).
6. B.D. Brooks, et al., Drug Discov. Today 19, 1040–1044 (2014).
7. P. Safsten, Methods Mol.Biol. 524, 67–76 (2009).
8. D. G. Myszka, Methods Enzymol. 323, 325–340 (2000).
9. R. Karlsson and A. Falt, J. Immunol. Methods 200, 121–133 (1997).
10. R. L. Rich, and D. G. Myszka, J. Mol. Recognit. 14, 273–294 (2001).
11. R. Karlsson, Biophysical Reviews 8 (4) 347–358 (2016).
12. A. S. Rathore, Trends in Biotechnology 27, 698–705 (2009).
13. N. Nupur, et al., mAbs 10 (1) 143–158 (2018).
14. M. Federici et al., Biologicals 41, 131–147 (2013).
15. FDA, Guidance for Industry: Scientific Considerations in Demonstrating Biosimilarity to a Reference Product (Rockville, MD, April 2015).
16. M. Schraeml and M. Biehl, Antib. Methods Protoc. 901, 171 –181 (2012).
17. P. Safsten, Methods Mol.Biol. 524, 67–76 (2009).
18. Å. Frostell, et al., Anal. Biochem. 477, 1–9 (2015).
19. Accurate comparability assessment of a biosimilar interferon in process development, Cytiva, Application Note 29-1154-78 AC (2014).
20. A. Moberg, et al., J. Pharm. Biomed. Anal. 78, 224–232 (2013).
21. D. G. Myszka, Methods Enzymol. 323, 325–340 (2000).
22. I. Navratilova, et al., Anal. Biochem. 355, 132–139 (2006).
23. T. Kitazawa et al., Nat. Med. 18 (10) 1570–1574 (2012).





