By Ramesh Kumar, Manidipa Banerjee, Anurag S. Rathore
Analytical and functional characterization of virus-like particles enables process reproducibility and product consistency.
Virus-like particles (VLPs) are viral capsid shells assembled without encapsulated nucleic acid. These particles resemble native virions, are highly ordered and repetitive, and are approximately 20 to 200 nm in diameter (1–3). VLPs, like native virions, can be non-enveloped or enveloped and icosahedral or pleomorphic (4–7). The uniformity and self-assembly of VLPs, along with their ability to withstand chemical modifications primarily on the outer surface, make them flexible and stable alternatives to nanoparticles, such as liposomes and metal assemblies (1,8). At least 110 VLPs have been constructed so far from 30 different virus families (1), and a variety of expression systems ranging from bacteria to mammalian cell systems have been used for their production (9). VLP surfaces have been modified through genetic or chemical means for display of molecules or antigens of interest (1,10), and manipulation of particles pre- and post-assembly has resulted in encapsulation of a variety of cargo molecules (3).
In this 44th article in the “Elements of Biopharmaceutical Production” series, the authors review the status of VLPs as therapeutics as well as challenges associated with their analytical and functional characterization. Concerns related to their stability are also discussed.
Applications of VLPs
The well-defined structure of VLPs, stability, non-infectious nature, and their ability to encapsulate molecules have resulted in tremendous applications of these particles in the areas of biotechnology, chemistry, and therapeutics (see Figure 1). Some of the therapeutic applications of VLPs are discussed as follows.
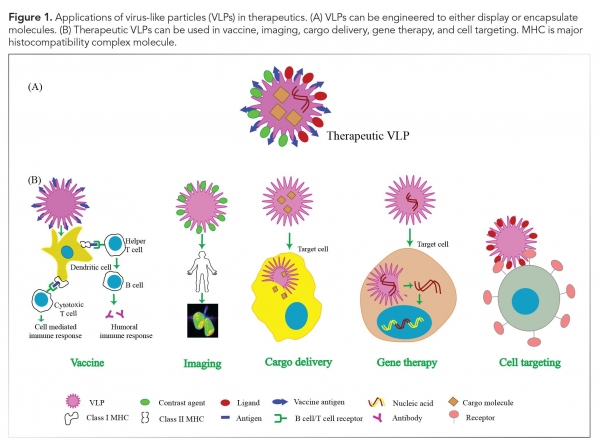
Vaccine development
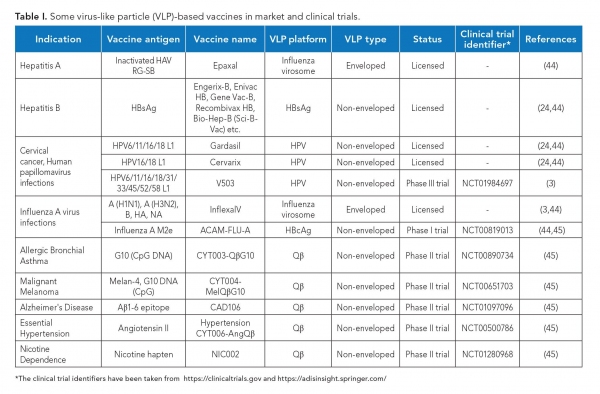
Traditional viral vaccines, which are based on inactivated or attenuated infectious virus, are difficult to produce and may cause adverse reactions in immunocompromised patients (9). Several VLPs can be easily produced at a large scale (4) and have a better safety profile due to the lack of viral genome. Further, owing to their highly repetitive surface structure and resemblance to native virions, these are highly immunogenic in nature. Inside the body, they are actively taken up by the antigen presenting cells (9) and induce strong humoral as well as cell-mediated immune responses, even without adjuvants (1). VLPs have thus emerged as an excellent alternative to traditional viral vaccines. Further, they can also be used as vaccine adjuvants (8). The first VLP-based vaccine was approved in 1986 (11), and since then, several such vaccines have been licensed and many more are in clinical trials (see Table I).
Bioimaging and diagnostics
VLPs are biocompatible, have low toxicity, and can be conjugated to fluorescent dyes, probes, or contrast agents to obtain better contrast with low toxicity in magnetic resonance imaging and positron emission tomography (6). This makes them excellent tools for in-vitro and in-vivo bioimaging as well as live cell imaging (3). Further, the potential of VLPs to target specific cell types increases signal-to-noise ratio in diagnostic imaging, thus providing better resolution (8). VLPs displaying specific antigenic proteins can also be used as diagnostic agents (1).
Delivery vehicles
VLPs derived from human viruses are conditioned to disassemble in the intracellular milieu and are therefore particularly well suited to deliver cargos within the body. VLPs have been shown to encapsulate a variety of cargo including foreign DNA, plasmid DNA, RNA (4), genome editing machinery, siRNA, proteins, and peptides for in-vitroand in-vivo applications. Further, drug molecules such as anticancer drugs, Adriamycin (doxorubicin hydrochloride), and aleomycin can also be loaded and delivered to target cell types (3,4). Encapsulation of drug molecules within VLPs, besides providing the advantage of targeted delivery, also prevents degradation of drug molecules in the bloodstream (3).
Gene therapy
The ability of VLPs to deliver specific genes make them attractive tools for gene therapy. Traditional tools for gene therapy, such as liposomes, have restricted packaging capacity, production difficulties, and undesirable immunological properties, whereas VLPs can potentially be engineered to overcome these limitations, thus opening a new paradigm for gene therapy (1).
Cell targeting
Cell type-specific ligands, such as proteins, aptamers, and small molecules, can be chemically or genetically coupled to the outer surface of VLPs to achieve cell-specific targeting, which can be further useful in medicine and research (3, 12). Multivalent display of such targeting moieties also assists specific localization of VLPs.
Besides these applications in therapeutics, VLPs are an excellent model system for studying the initial host interaction pathways of biosafety level 4 restricted viruses and the modes of virus assembly (4). They have also found use in numerous other fields including biocatalysis, energy production, development of nanomaterials (6), antibody purification (1), antigen screening (3), and tissue engineering (13).
Characterization of VLPs
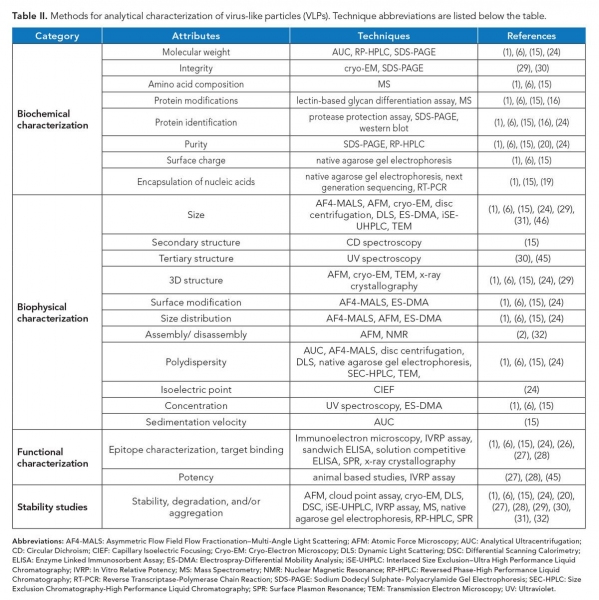
Post-production characterization of VLPs in terms of composition, morphology, homogeneity, and stability is essential prior to their usage (see Figure 2). Various tools are available in this regard as discussed as follows and summarized in Table II. For simplicity, the analysis can be categorized into compositional, morphological, functional, and stability based.
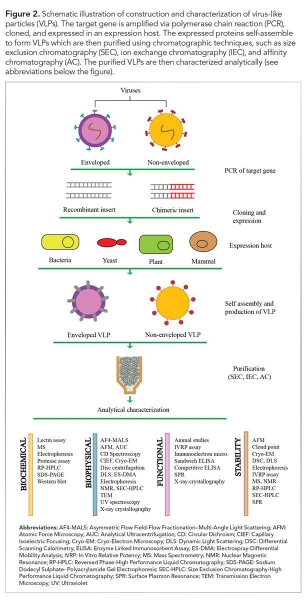
VLP composition analysis
Depending on the parent virus, VLPs may be composed of one or multiple proteins, and may contain a lipid layer. In addition, various non-desired impurities such as media components, debris, host DNA or RNA, and exosomes may be retained in the final product (14). Modification of VLPs may also affect their downstream application. Identification of the composition is thus a prerequisite for characterization of VLPs.
Mass spectrometry (MS) is a robust tool to analyze the molecular mass, amino acid composition, and post-translational modifications (PTMs) of the capsid proteins present in VLPs. The high sensitivity of this technique facilitates detection of even single amino acid changes (15). PTMs such as glycosylation can also be analyzed using lectin-based glycan differentiation assay (16). Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) can be used to analyze the molecular weight and protein purity of VLPs. The identity of individual capsid proteins can be confirmed using Western blotting in conjunction with SDS–PAGE (1).
Reverse phase–high-performance liquid chromatography (RP–HPLC) is a frequently used analytical tool for protein characterization and purification, and has been recently shown to be useful for characterization of VLPs. It can also be used to analyze molecular mass and VLP disassembly (17).
Virus capsid proteins that self-assemble to generate particles sometimes contain flexible, positively charged regions that associate with viral genome. In absence of cognate genome, and for charge neutralization prior to assembly, particles encapsulate a variety of nucleic acids from the expression host. Some of the packaged RNA can even be transposable elements (18). Also, expression host DNA frequently co-purify with VLPs. VLPs as a delivery system for nucleic acids require encapsulation of DNA or RNA. To check the association or encapsulation of nucleic acid with VLPs, native agarose gel electrophoresis can be performed (15). The type of nucleic acid encapsulated, whether DNA or RNA, can also be analyzed by treating the VLP sample with respective nuclease and then analyzing on the agarose gel. Loss of bands upon nuclease treatment suggests the encapsulation of respective nucleic acids. The sequence of encapsulated nucleic acids within VLPs can also be confirmed via reverse transcriptase polymerase chain reaction (19) and next-generation sequencing (1).
Morphology analysis
Correct size of the particles is a critical attribute of a VLP preparation. Various analytical tools can be used to analyze the size of VLPs, such as electrospray-differential mobility analysis (ES–DMA), asymmetrical flow field-flow fractionation with multiple angle light scattering (AF4–MALS), dynamic light scattering (DLS), disc centrifugation, size-exclusion chromatography (SEC), analytical ultracentrifugation (AUC) (15), and nanoparticle tracking analysis (NTA) (20).
ES–DMA quantifies the external size of VLPs based on their electrical mobility, with resolution in the sub-nanometer range. Such high resolution allows analysis of even small shifts in particle size and hence can be used to analyze size distribution in a VLP preparation (21). Physical parameters of VLPs such as molecular mass, density, size distribution, polydispersity, and conformation can be studied at high resolution, even for a heterogeneous sample without causing any aggregation using an AF4-MALS technique (22). DLS, disc centrifugation, AUC (15), SEC, and NTA (20) are other tools that can be used to check the size and polydispersity of VLPs. AUC calculates the sedimentation velocity of VLPs and provides information about the molecular weight, size distribution, and conformation of the particles (15). A SEC profile of a VLP is indicative of its structure, and any variation in it indicates possible variation in the structure of a VLP (23). NTA offers a non-invasive method to quickly analyze size distribution, identify aggregates, and calculate the concentration of particles (20). It is to be noted that enveloped, pleomorphic VLPs typically offer a range of sizes and shapes and are relatively difficult to characterize compared to non-enveloped ones. For the former, frequently, membrane vesicles decorated with surface proteins co-purify with VLPs, which can be identified using high resolution structural methods, such as cryo-electron microscopy (cryo-EM).
The structural characteristics of VLPs can be mapped by various techniques including transmission electron microscopy, cryo-EM, atomic force microscopy (AFM), and x-ray crystallography, the latter being applicable to uniform, non-pleomorphic particles. X-ray crystallography and cryo-EM can be used to generate three-dimensional structures of a uniform population of VLPs at high resolution and reveal the precise location of epitopes. These methods can also be used to study morphology and size distribution of particles (24). Spectroscopic techniques like ultraviolet spectroscopy, circular dichroism spectroscopy, and intrinsic and extrinsic fluorescence can be used to study the secondary and tertiary structure of VLPs and their conformational changes (25).
Functional analysis
Functional characterization of VLPs includes assays to evaluate binding ability of drug delivery vehicles to specific target cells, or antigenicity and immunogenicity of vaccine candidates. These assays are crucial for ensuring correct downstream application of VLPs and determining the potency of the preparation. Both in-vivo studies in animal models and in-vitro studies using methods such as enzyme linked immunosorbent assay (ELISA), surface plasmon resonance (SPR), and in-vitro relative potency (IVRP) assays can be performed in this regard (24). Solution competitive ELISA tracks the correct display of epitopes on VLPs with a neutralizing monoclonal antibody against the respective epitope, which allows real-time monitoring of relative antigenicity of VLPs during manufacturing as well as in product form. Also, the technique can be used to detect alterations in epitopes, if any, such as during storage. SPR offers a high-throughput, rapid, label-free method to detect association of VLPs with their targets in real time (26).
An IVRP assay reports the qualitative and quantitative epitope composition of a VLP sample and is a good predictor of VLP potency. The results of the assay have been found to correlate well with human clinical data (27). It is a good alternative to animal potency assays.
Stability analysis
The stability of VLPs varies based on the composition, size, presence of lipids, and other characteristics. The shape, size, and arrangement of coat proteins on stable particles may get altered in response to stress conditions, such as pH. For drug delivery vehicles, identification of conditions that favor their disassembly and reassembly is essential for their optimal packaging and eventual release of cargo molecules. Thermostability of VLPs can be evaluated by differential scanning calorimetry (DSC) and cloud point assays. While DSC is used to detect unfolding of component proteins, cloud point assays reveal the tendency of VLPs to aggregate during conditions of thermal stress. Further, aggregation and degradation of VLPs can be studied by AFM, cryo-EM, DLS, interlaced size exclusion–ultra high performance liquid chromatography, IVRP assay, MS, native agarose gel electrophoresis, RP–HPLC, and SPR (1,6,15,20,24,27–32).
These biophysical, biochemical, and immunochemical methods ensure process reproducibility and product consistency in recombinant VLPs during the manufacturing process. These characterization methods are thus key to preclinical and clinical studies, as well as commercial production of VLPs.
Recent developments
Various breakthroughs with respect to VLP production, characterization, and their applications have been reported in the past few years, such as the following:
- An SEC-based VLP purification strategy, for large-scale production of VLP-based vaccines, and an size-exclusion high performance liquid chromatography method have been developed as efficient methods for analysis and quality checks of VLPs. These allow quick analysis of VLP assembly for monitoring consistency in VLP formulations during production
and storage (23). - A 19F-nuclear magnetic resonance-based system for studying VLP disassembly has been established. Studying disassembly of VLPs
is important especially when
VLPs have to be used as carriers, which are dependent on their breakdown (2). - A rapid method for characterization of VLPs via interlaced SEC has been reported. With this advancement, the time of analysis has been reduced from 30–60 minutes per sample to 3.1 minutes (31).
- Investigation of spectroscopic characteristics of VLPs and generation of spectral fingerprints have been proposed. Generation of spectral libraries of VLPs would allow for spectroscopy-based detection of VLPs (33).
- For high-throughput screening of formulations with small amounts of sample, a DLS-AF4 based method has been developed (15,34).
- A VLP-based vaccine for malaria, Mosquirix (RTS/S), has received approval from regulatory authorities. This is the first vaccine for malaria in the world (9).
- Attempts have been taken to widen the spectrum coverage of VLP-based vaccine using chimeric systems, such as by adding a new epitope on existing VLP platforms (24).
- Development of a VLP-based vaccine and diagnostic assay has been reported for Zika virus (35).
- VLPs have been used as biocatalysts and biosensors (36).
- Catalytic nanomaterials have been developed for label-free detection of VLPs (37).
- Capillary electrophoresis-based methods have been developed for characterization of VLPs (38,39).
Challenges encountered in development of therapeutic VLPs
While VLPs have established themselves as promising candidates for therapeutics and other applications, there are limitations that hinder their development. The time involved in production, purification, and characterization is currently a major challenge in developing VLP-based products (3, 40, 41). VLPs based on enveloped, pleomorphic viruses or those containing multiple proteins with extensive PTMs are difficult to generate (12, 42). Most of the methods or techniques used for their characterization, such as AF4, are complicated or not readily available (22). The stability of VLPs, particularly lipid-containing VLPs, during formulation and storage are other challenges in the use of VLP-based therapeutics (34, 42). Repeated administration of VLPs may lead to inflammation and other responses, while incorporation of transposable elements may lead to other complications (18, 43).
Future perspectives
Since the approval of the first VLP-based vaccine in 1986, the application of VLPs in therapeutics has grown exponentially. It is expected that future studies will be aimed at developing technologies that lead to large-scale yet economical production of VLPs and establishment of an appropriate analytical platform for rapid characterization. Improvement in methods may include development of in-silico tools to predict surface epitopes for VLP design, development of spectral libraries for rapid identification, and development of more efficient and rapid SEC columns. A complete understanding of the distribution of VLPs in the human body and toxicity and immunogenicity profiling are other essential topics for VLP-related future research. It is hoped that fast and efficient production methods and efficient analytical platforms may lead to more extensive usage of VLPs as potential therapeutics.
References
- A. Zeltins, Mol. Biotechnol. 53 (1) 92–107 (2013).
- R. L. C. Leung et al., J. Am. Chem. Soc. 139 (15) 5277–5280 (2017).
- Z. Shirbaghaee and A. Bolhassani, Biopolymers 105 (3) 113–132 (2016).
- D. Yan, et al., Appl. Microbiol. Biotechnol. 99 (24) 10415–10432 (2015).
- W. Akahata et al., Nat. Med. 16 (3) 334–338 (2004).
- X. Ding et al., Biotechnol. J. 13 (5) e1700324 (2018).
- S. Paola Sanchez-Rodriguez, L. Munch-Anguiano, and I. Bustos-Jaimes, Curr. Chem. Biol. 4 (3) 231–243 (2010).
- M. J. Rohovie, M. Nagasawa, and J. R. Swartz, Bioeng. Transl. Med. 2 (1) 43–57 (2017).
- M. O. Mohsen et al., Semin. Immunol. 34, 123–132 (2017).
- J. G. Heddle, S. Chakraborti, and K. Iwasaki, Curr. Opin. Struct. Biol. 43, 148–155 (2017).
- J. Chroboczek, I. Szurgot, and E. Szolajska, Mol. Biotechnol. 61 (2) 531–539 (2014).
- P. Pushko, P. Pumpens, and E. Grens, Intervirology 56 (3) 141–165 (2013).
- A. M. Wen and N. F. Steinmetz, Chem. Soc. Rev. 45 (15) 4074–4126 (2016).
- P. Kramberger, L. Urbas, and A. Štrancar, Hum. Vaccines Immunother. 11 (4) 1010–1021 (2015).
- L. H. L. Lua et al., Biotechnol. Bioeng. 111 (3) 425–440 (2014).
- L. M. Branco et al., Virol. J. 7, 279 (2010).
- A. Shytuhina et al., J. Chromatogr. A 1364, 192–197 (2014).
- A. Routh, T. Domitrovic, and J. E. Johnson, Proc. Natl. Acad. Sci. USA 109 (6) 1907–1912 (2012).
- J. P. Phelps et al., J. Biotechnol. 128 (2) 290–296 (2007).
- P. Steppert et al., J. Chromatogr. A 1487, 89–99 (2017).
- L. F. Pease et al., Biotechnol. Bioeng. 102 (3) 845–855 (2009).
- M. Wagner et al., Anal. Chem. 86(11), 5201–5210 (2014).
- Y. Yang et al., Vaccine 33(9), 1143–1150 (2015).
- Q. Zhao et al., Trends Biotechnol. 31(11), 654–663 (2013).
- N. K. Jain et al., Adv. Drug Deliv. Rev. 93, 42–55 (2015).
- A. M. Mulder et al., PLoS One 7(4) , e33235 (2012).
- M. Shank-Retzlaff et al., Hum. Vaccin. 1 (5) 191–197 (2005).
- M. L. Shank-Retzlaff et al., Hum. Vaccin. 2 (4) 147–154 (2006).
- Q. J. Zhao et al., Hum. Vaccin. Immunother. 10 (3) 734–739 (2014).
- X. Zhang et al., Vaccine 32 (32) 4039–4050 (2014).
- C. Ladd Effio, S. A. Oelmeier, and J. Hubbuch, Vaccine 34 (10) 1259–1267 (2016).
- Q. Zhao et al., Nanomedicine Nanotechnology, Biol. Med. 8 (7) 1182–1189 (2012).
- P. J. Kervalishvili and T. N. Bzhalava, Am. J. Condens. Matter Phys. 6 (1) 7–16 (2016).
- J. Mohr et al., Methods 60 (3) 248–256 (2013).
- H. Garg et al., J. Virol. 91 (20) e00834-17 (2017).
- M. Raeeszadeh-Sarmazdeh et al., Curr. Opin. Chem. Eng. 13, 109–118 (2016).
- S. Sykora et al., ChemBioChem 18 (11) 996–1000 (2017).
- V. Bettonville et al., Talanta 175, 325–330 (2017).
- D. Gollapudi et al., Electrophoresis 38 (20) 2610–2621 (2017).
- H. K. Charlton Hume et al., Biotechnol. Bioeng. 116 (4) 919–935 (2019).
- J. Zepeda-Cervantes, J. O. Ramírez-Jarquín, and L. Vaca, Front. Immunol. 11, 1100 (2020).
- S. Dai, H. Wang, and F. Deng, J. Immunol. Sci. 2 (2) 36–41 (2018).
- B. Schwarz and T. Douglas, WIREs Nanomed Nanobiotechnol 7 (5) 722–735 (2015).
- N. Kushnir, S. J. Streatfield, and V. Yusibov, Vaccine 31 (1) 58–83 (2012).
- X. Huang et al., npj Vaccines 2 (3) 1–8 (2017).
- L. F. Pease III, Trends Biotechnol. 30 (4) 216–223 (2012).
About the authors
*To whom all correspondence should be addressed.
Ramesh Kumar is a graduate student, Manidipa Banerjee is a professor in the Kusuma School of Biological Sciences, and Anurag S. Rathore*, PhD, is professor in the Department of Chemical Engineering and coordinator, DBT Center of Excellence for Biopharmaceutical Technology, all at the Indian Institute of Technology, Delhi, Hauz Khas, New Delhi, 110016, India, asrathore@biotechcmz.com, Phone: +91-11-26591098.